Library
RNA Structure, types, and functions- a quick revision
- February 1, 2020
- Posted by: Namrata Chhabra
- Category: Learning resources Lecture notes Lecture notes Lecture notes Library Molecular Biology Molecular Biology
RNA (Ribonucleic acid )
Ribonucleic acid (RNA) is a polymer of purine and pyrimidine ribonucleotides linked together by 3′,5′-phosphodiester bridges analogous to those in DNA (Figure-1).
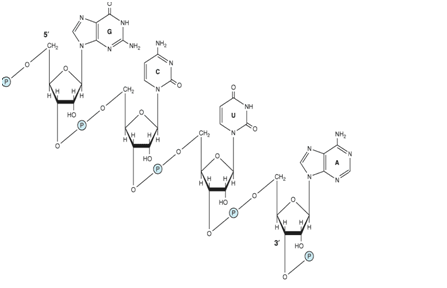
Figure 1- Showing an RNA fragment, the ribonucleotides are linked together by 3′-5′ phosphodiester linkages. Ribose is the principal sugar in RNA
Differences between RNA and DNA
Although sharing many features with DNA, RNA possesses several specific differences:
| S.No. | RNA | DNA |
| 1) | Single-stranded mainly except when self-complementary sequences are there, it forms a double-stranded structure (Hairpin structure) | Double-stranded (Except for certain viral DNA s which are single-stranded) |
| 2) | Ribose is the main sugar | The sugar moiety is deoxyribose |
| 3) | Pyrimidine components differ. Thymine is never found (Except-tRNA) | Thymine is always there, but uracil is never found |
| 4) | Being a single-stranded structure- It does not follow Chargaff’s rule | It does follow Chargaff’s rule. The total purine content in a double-stranded DNA is always equal to pyrimidine content. |
| 5) | RNA can be easily destroyed by alkalies to cyclic diesters of mononucleotides. | DNA resists alkali action due to the absence of the OH group at the 2’ position |
| 6) | RNA is a relatively a labile molecule that undergoes easy and spontaneous degradation | DNA is a stable molecule. The spontaneous degradation is very slow. The genetic information can be stored for years together without any change. |
| 7) | Mainly cytoplasmic, but also present in the nucleus (primary transcript and small nuclear RNA) | Mainly found in the nucleus, extranuclear DNA is found in mitochondria, and plasmids, etc. |
| 8) | The base content varies from 100- 5000. The size is variable. | Millions of base pairs are there depending upon the organism |
| 9) | There are various types of RNA – mRNA, rRNA, tRNA, snRNA, siRNA, miRNA, and hnRNA. These RNAs perform different and specific functions. | DNA is always of one type and performs the function of storage and transfer of genetic information. |
| 10) | No variable physiological forms of RNA are found. The different types of RNA do not change their forms | There are various forms of DNA (A to E and Z) |
| 11) | RNA is synthesized from DNA; it cannot form DNA(except by the action of reverse transcriptase). It cannot duplicate (except in certain viruses where it is a genomic material ) | DNA can form DNA by replication, and RNA can be formed by transcription. |
| 12) | Many copies of RNA are present per cell | A single copy of DNA is present per cell. |
Types of RNA
In all prokaryotic and eukaryotic organisms, three main classes of RNA molecules exist-
1) Messenger RNA (m- RNA)
2) Transfer RNA (t- RNA)
3) Ribosomal RNA (r- RNA)
The others are –
- Small nuclear RNA (snRNA)
- Micro RNA(mi RNA)
- Small interfering RNA(Si RNA) and
- Heterogeneous nuclear RNA (hnRNA).
1) Messenger RNA (mRNA
- Comprises only 5% of the RNA in the cell
- Most heterogeneous in size and base sequence
- All members of the class function as messengers carrying the information in a gene to the protein-synthesizing machinery
Structural Characteristics of mRNA
- The 5’ terminal end is capped by a 7-methyl Guanosine triphosphate cap.
- The cap is involved in the recognition of mRNA by the translating machinery.
- It stabilizes m- RNA by protecting it from 5’ exonuclease
- The 3’end of most mRNAs have a polymer of Adenylate residues( 20-250)
- The tail prevents the attack by 3’ exonucleases. Histones and interferons do not contain poly-A tails
- On both 5’ and 3’ end, there are noncoding sequences which are not translated (NCS)
- The intervening region between noncoding sequences present between 5’ and 3’ end is called the coding region. This region encodes for the synthesis of a protein.
- The mRNA molecules are formed with the help of the DNA template during the process of transcription.
- The sequence of nucleotides in m RNA is complementary to the sequence of nucleotides on template DNA.
- The sequence carried on mRNA is read in the form of codons.
- A codon is made up of 3 nucleotides
- The mRNA is formed after processing of heterogeneous nuclear RNA
2) Heterogeneous nuclear RNA (hnRNA)
- In mammalian nuclei, hnRNA is the immediate product of gene transcription
- The nuclear product is heterogeneous in size (Variable) and is very large.
- The molecular weight may be more than 107, while the molecular weight of m RNA is less than 2x 106
- 75 % of hnRNA is degraded in the nucleus, and only 25% is processed to mature m RNA
3) Transfer RNA (tRNA)
- Transfer RNA is the smallest of three major species of RNA molecules
- They have 74-95 nucleotide residues
- They are synthesized by the nuclear processing of a precursor molecule
- They transfer the amino acids from the cytoplasm to the protein-synthesizing machinery, hence the name t RNA.
- They are easily soluble, hence called “Soluble RNA or s RNA
- They are also called Adapter molecules since they act as adapters for the translation of the sequence of nucleotides of the m RNA into specific amino acids
- There are at least 20 species of tRNA one corresponding to each of the 20 amino acids required for protein synthesis.
Structural characteristics of tRNA
A) Primary structure- The nucleotide sequence of all the t RNA molecules allows extensive intrastrand complementarity that generates a secondary structure.
B) Secondary structure– Every single tRNA shows extensive internal base-pairing and acquires a clover leaf-like structure. The structure is stabilized by hydrogen bonding between the bases and is a consistent feature.
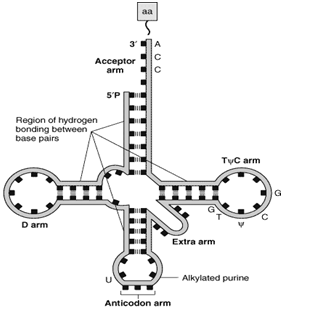
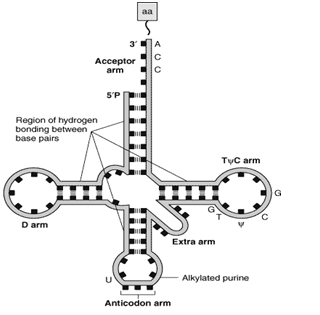
Cloverleaf structure (Figure-2(a)
All t-RNA contain five main arms or loops, which are as follows-
a) Acceptor arm
b) Anticodon arm
c) D HU arm
d) TΨ C arm
e) Extra arm
a) Acceptor arm
- The acceptor arm is at 3’ end
- It has seven base pairs
- The end sequence is unpaired Cytosine, Cytosine-Adenine at the 3’ end
- The 3’ OH group terminal of Adenine binds with the carboxyl group of amino acids
- The t RNA bound with amino acid is called Amino acyl t RNA
- CCA attachment is done post-transcriptionally,
b) Anticodon arm
- Lies at the opposite end of acceptor arm
- 5 base pairs long
- Recognizes the triplet codon present in the m RNA
- The base sequence of the anticodon arm is complementary to the base sequence of the m RNA codon.
- Due to complementarity, it can bind specifically with m RNA by hydrogen bonds.
c) DHU arm
- It has 3-4 base pairs
- It serves as the recognition site for the enzyme (aminoacyl t RNA synthetase) that adds the amino acid to the acceptor’s arm.
d) TΨC arm
- This arm is opposite the DHU arm
- Since it contains pseudouridine, that is why it is so-named
- It is involved in the binding of t RNA to the ribosomes
e) Extra arm or Variable arm
- About 75 % of t RNA molecules possess a short extra arm
- If about 3-5 base pairs are present, the t-RNA is said to be belonging to class 1. The majority of tRNA belongs to class 1.
- The tRNA belonging to class 2 has a long extra arm and 13-21 base pairs in length.
C) The tertiary structure of t- RNA- Figure-2(b)
- The L-shaped tertiary structure is formed by further folding of the cloverleaf due to hydrogen bonds between T and D arms.
- The base-paired double-helical stems get arranged into two double-helical columns, continuous and perpendicular to one another.
(a)
(b) 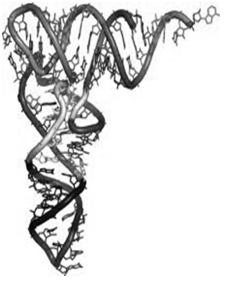
Figure-2-(a) Showing secondary (cloverleaf ) structure of t-RNA. The carboxyl group of amino acid is attached to the 3’OH group of the Adenine nucleotide of the acceptor arm. The anticodon arm base pairs with the codon present on the mRNA. (b) The tertiary structure of t RNA is formed by further folding of the secondary structure of t RNA.
4) Ribosomal RNA (rRNA)
The mammalian ribosome contains two major nucleoprotein subunits—a larger one with a molecular weight of 2.8 x 106 (the 60S) and a smaller subunit with a molecular weight of 1.4 x 106 (40S) (Figure 3).
- The 60S subunit contains a 5S ribosomal RNA (rRNA), a 5.8S rRNA, and a 28S rRNA; there are also probably more than 50 specific polypeptides.
- The 40S subunit is smaller and contains a single 18S rRNA and approximately 30 distinct polypeptide chains.
- All of the ribosomal RNA molecules except the 5S rRNA are processed from a single 45S precursor RNA molecule in the nucleolus. 5S rRNA is independently transcribed.
Functions of the ribosomal RNA
The functions of the ribosomal RNA molecules in the ribosomal particle are not fully understood, but they are necessary for ribosomal assembly and seem to play key roles in the binding of mRNA to ribosomes and its translation. Recent studies suggest that an rRNA component performs the peptidyl transferase activity and thus is an enzyme (a ribozyme).
5) Small RNAs
- Most of these molecules are complexed with proteins to form ribonucleoproteins and are distributed in the nucleus, in the cytoplasm, or in both.
- They range in size from 20 to 300 nucleotides and are present in 100,000–1,000,000 copies per cell.
Small Nuclear RNAs (snRNAs)
- snRNAs, a subset of the small RNAs, are significantly involved in mRNA processing and gene regulation
- Of the several snRNAs, U1, U2, U4, U5, and U6 are involved in intron removal and the processing of hnRNA into mRNA
- The U7 snRNA is involved in the production of the correct 3′ ends of histone mRNA—which lacks a poly(A) tail.
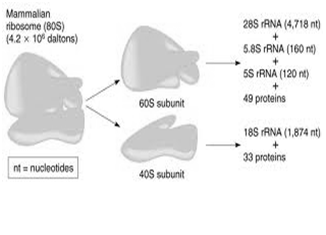
Figure 3- showing the structure of mammalian ribosomes- Ribosomal RNA is the structural component of ribosomes.
Micro RNAs, miRNAs, and Small Interfering RNAs, siRNAs
- These two classes of RNAs represent a subset of small RNAs; both play important roles in gene regulation.
- miRNAs and siRNAs cause inhibition of gene expression by decreasing specific protein production, albeit apparently via distinct mechanisms
Micro RNAs (miRNAs)
- miRNAs are typically 21–25 nucleotides in length and are generated by nucleolytic processing of the products of distinct genes/transcription units
- The small processed mature miRNAs typically hybridize via the formation of imperfect RNA-RNA duplexes within the 3′-untranslated regions of specific target mRNAs, leading via unknown mechanisms to translation arrest.
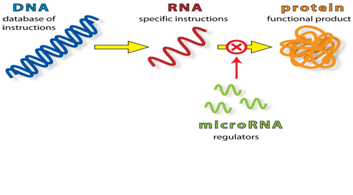
Figure 4- showing the mechanism of action of micro RNAs, micro RNAs, short non-coding RNAs present in all living organisms, have been shown to regulate the expression of at least half of all human genes. These single-stranded RNAs exert their regulatory action by binding messenger RNAs and preventing their translation into proteins.
Small Interfering RNAs (siRNAs)
- siRNAs are derived by the specific nucleolytic cleavage of larger, double-stranded RNAs to again form small 21–25 nucleotide-long products.
- These short siRNAs usually form perfect RNA-RNA hybrids with their distinct targets potentially anywhere within the length of the mRNA where the complementary sequence exists.
- Formation of such RNA-RNA duplexes between siRNA and mRNA results in reduced specific protein production because the siRNA-mRNA complexes are degraded by dedicated nucleolytic machinery.
Significance of mi RNAs and si RNAs
- Both miRNAs and siRNAs represent exciting new potential targets for therapeutic drug development in humans.
- In addition, siRNAs are frequently used to decrease or “knock-down” specific protein levels in experimental procedures in the laboratory, an extremely useful and powerful alternative to gene-knockout technology.
Author:Namrata Chhabra
Leave a Reply Cancel reply
You must be logged in to post a comment.
